Assessment of protein model structure accuracy estimation in CASP14: Old and new challenges - Kwon - 2021 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library

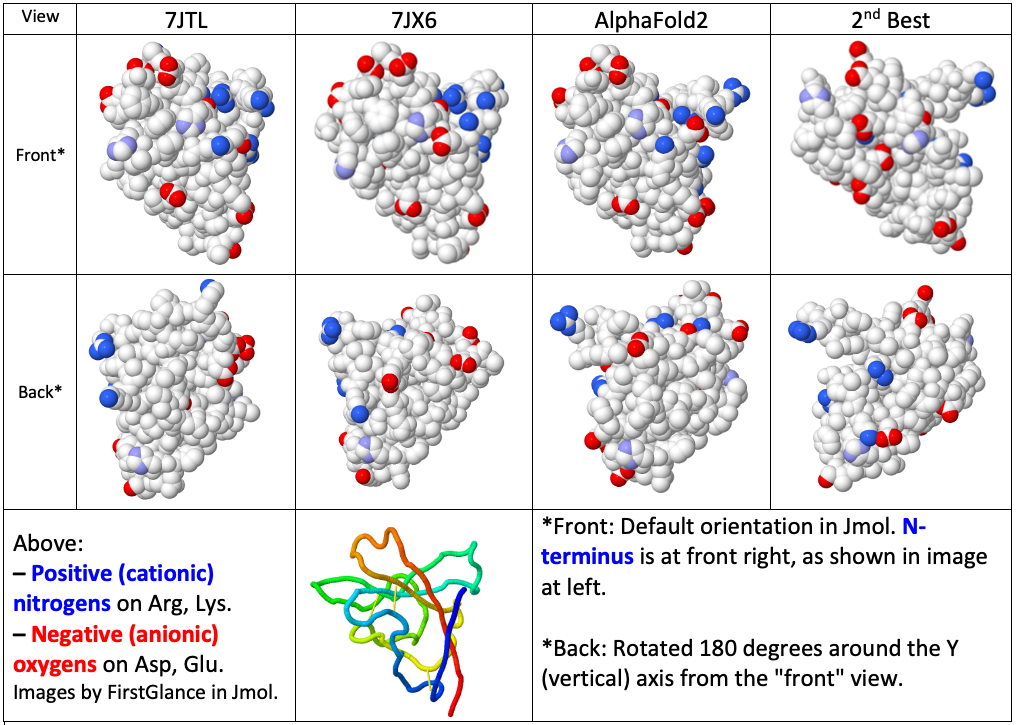
CASP14: what Google DeepMind's AlphaFold 2 really achieved, and what it means for protein folding, biology and bioinformatics | Oxford Protein Informatics Group

Global versus Local Hubs in Human Protein–Protein Interaction Network | Journal of Proteome Research

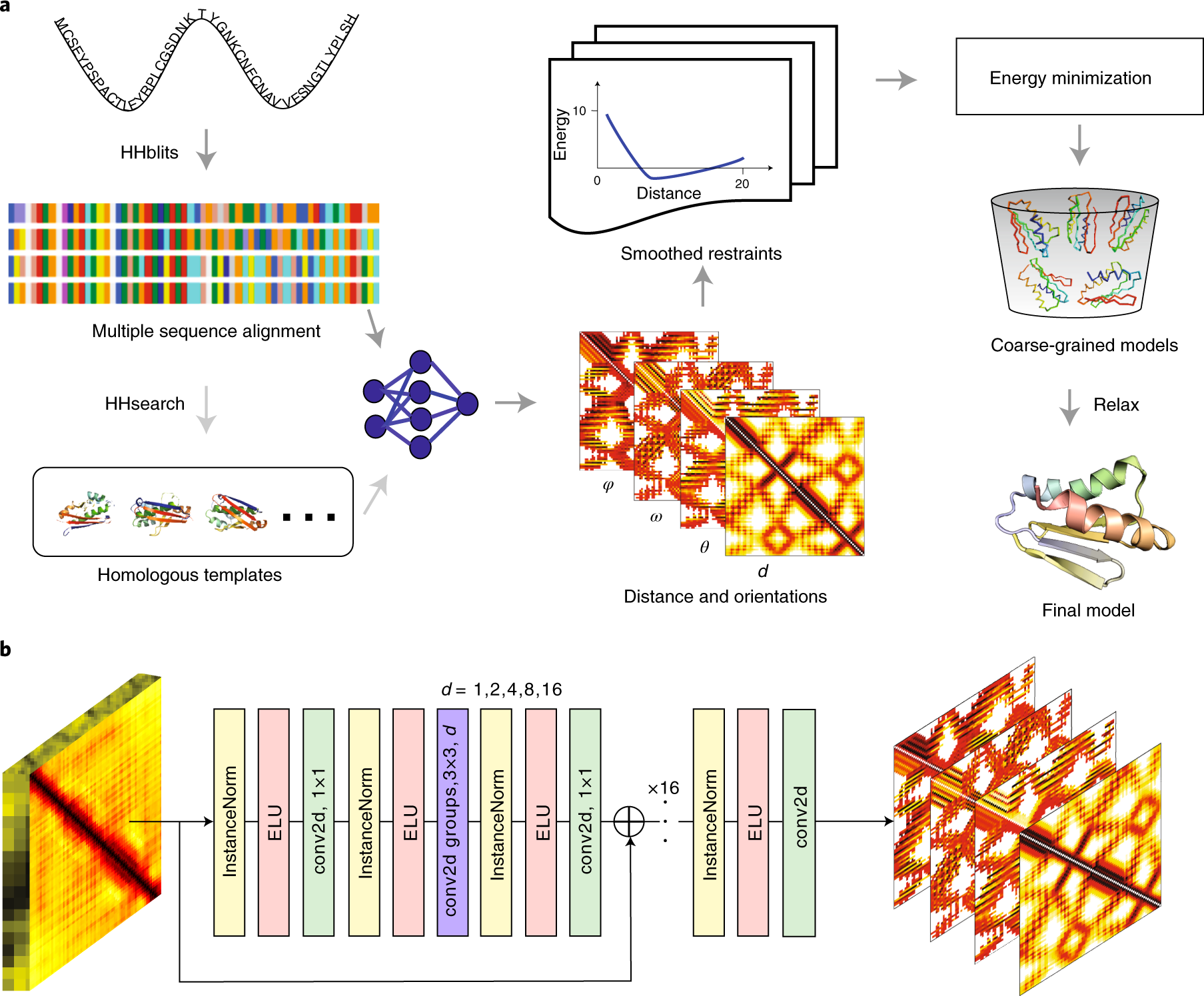
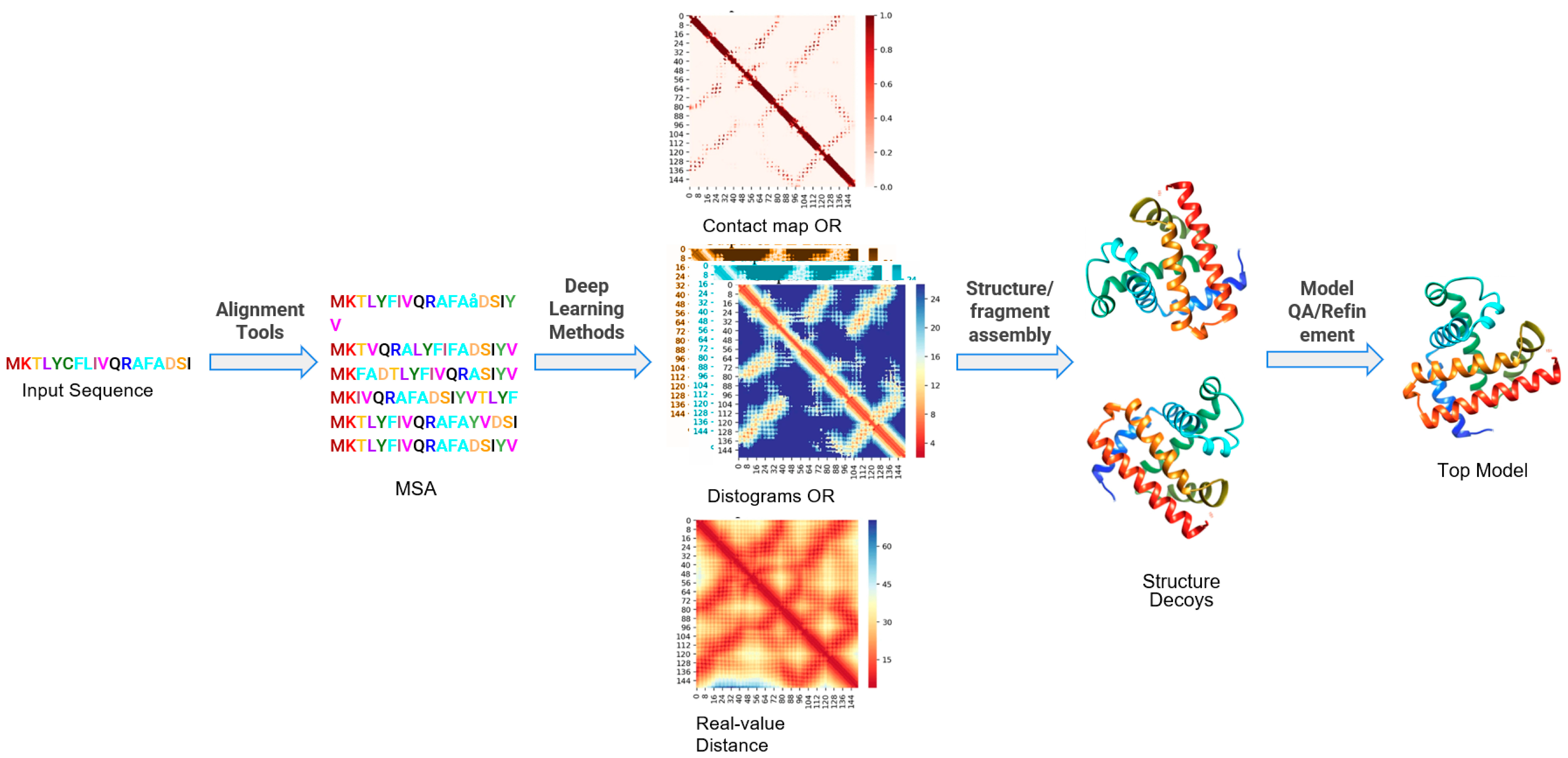
Protein tertiary structure modeling driven by deep learning and contact distance prediction in CASP13 | bioRxiv

Critical assessment of methods of protein structure prediction (CASP)—Round XII - Moult - 2018 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library
1 Protein structure Prediction. 2 Copyright notice Many of the images in this power point presentation are from Bioinformatics and Functional Genomics. - ppt download
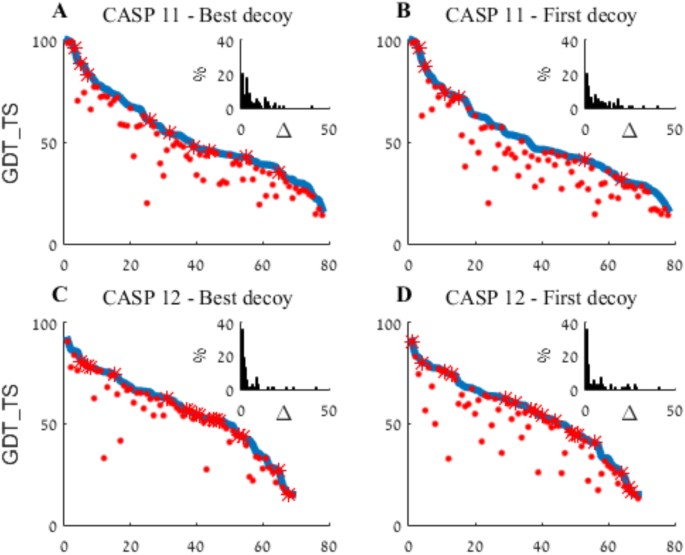
An analysis and evaluation of the WeFold collaborative for protein structure prediction and its pipelines in CASP11 and CASP12 | Scientific Reports

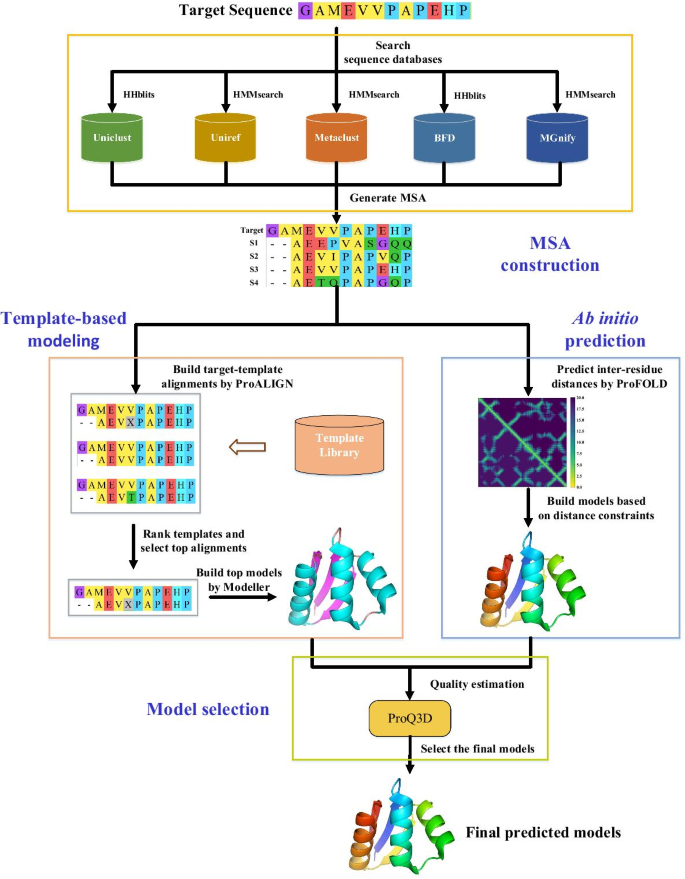
Improving protein tertiary structure prediction by deep learning and distance prediction in CASP14 - Liu - 2022 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library

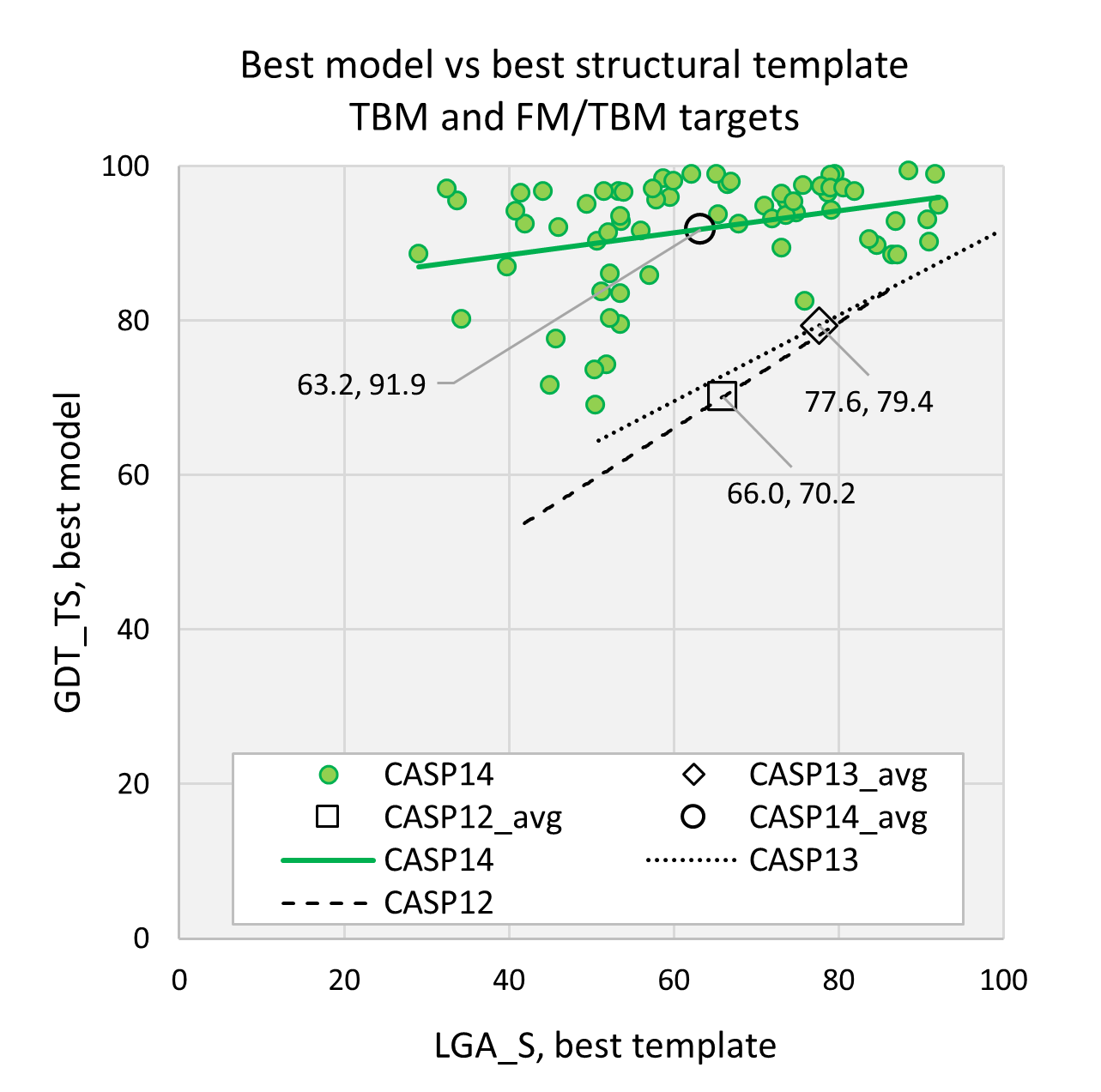
Topology evaluation of models for difficult targets in the 14th round of the critical assessment of protein structure prediction (CASP14) - Kinch - 2021 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library

TopModel: Template-Based Protein Structure Prediction at Low Sequence Identity Using Top-Down Consensus and Deep Neural Networks | Journal of Chemical Theory and Computation

ROPIUS0: A deep learning-based protocol for protein structure prediction and model selection and its performance in CASP14 | bioRxiv
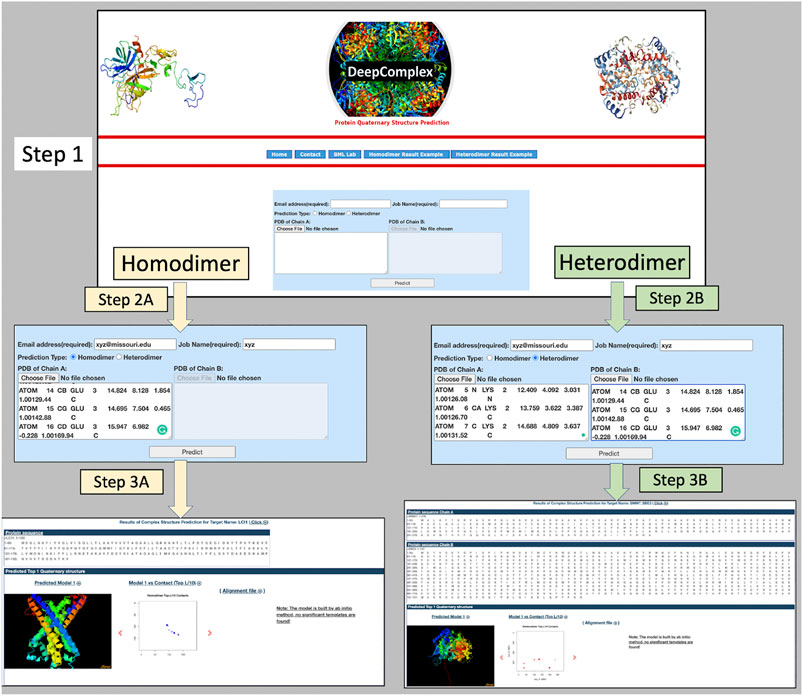
Frontiers | DeepComplex: A Web Server of Predicting Protein Complex Structures by Deep Learning Inter-chain Contact Prediction and Distance-Based Modelling

Topology evaluation of models for difficult targets in the 14th round of the critical assessment of protein structure prediction (CASP14) - Kinch - 2021 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library
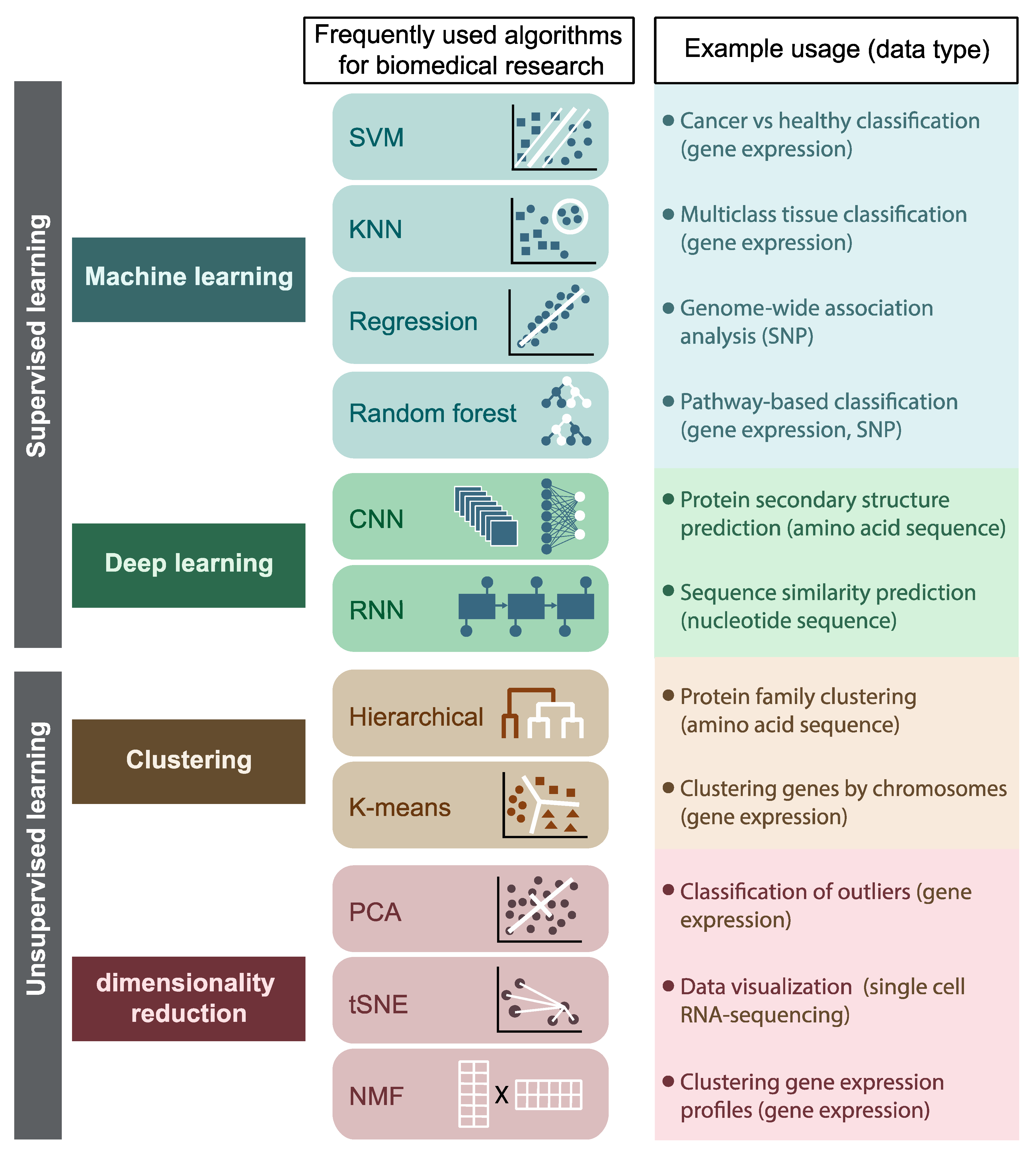
IJMS | Free Full-Text | Incorporating Machine Learning into Established Bioinformatics Frameworks | HTML

Assessment of protein model structure accuracy estimation in CASP13: Challenges in the era of deep learning - Won - 2019 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library

Ranking of the groups registered for the CASP14 season in the tertiary... | Download Scientific Diagram