CHARMM Force-Field Parameters for Morphine, Heroin, and Oliceridine, and Conformational Dynamics of Opioid Drugs | Journal of Chemical Information and Modeling

CHARMM‐GUI high‐throughput simulator for efficient evaluation of protein–ligand interactions with different force fields - Guterres - 2022 - Protein Science - Wiley Online Library

Relaxation of backbone bond geometry improves protein energy landscape modeling - Conway - 2014 - Protein Science - Wiley Online Library

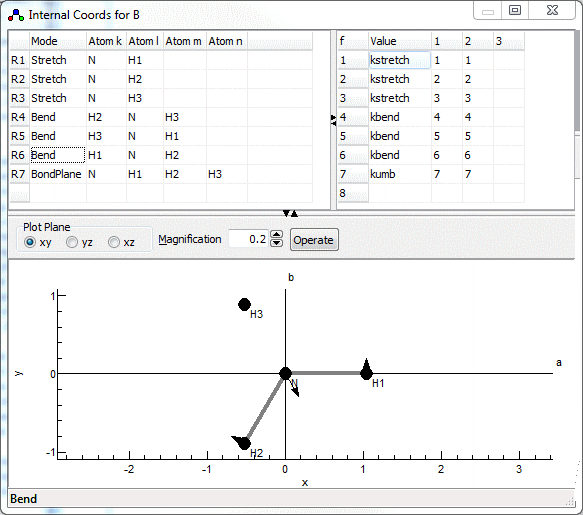
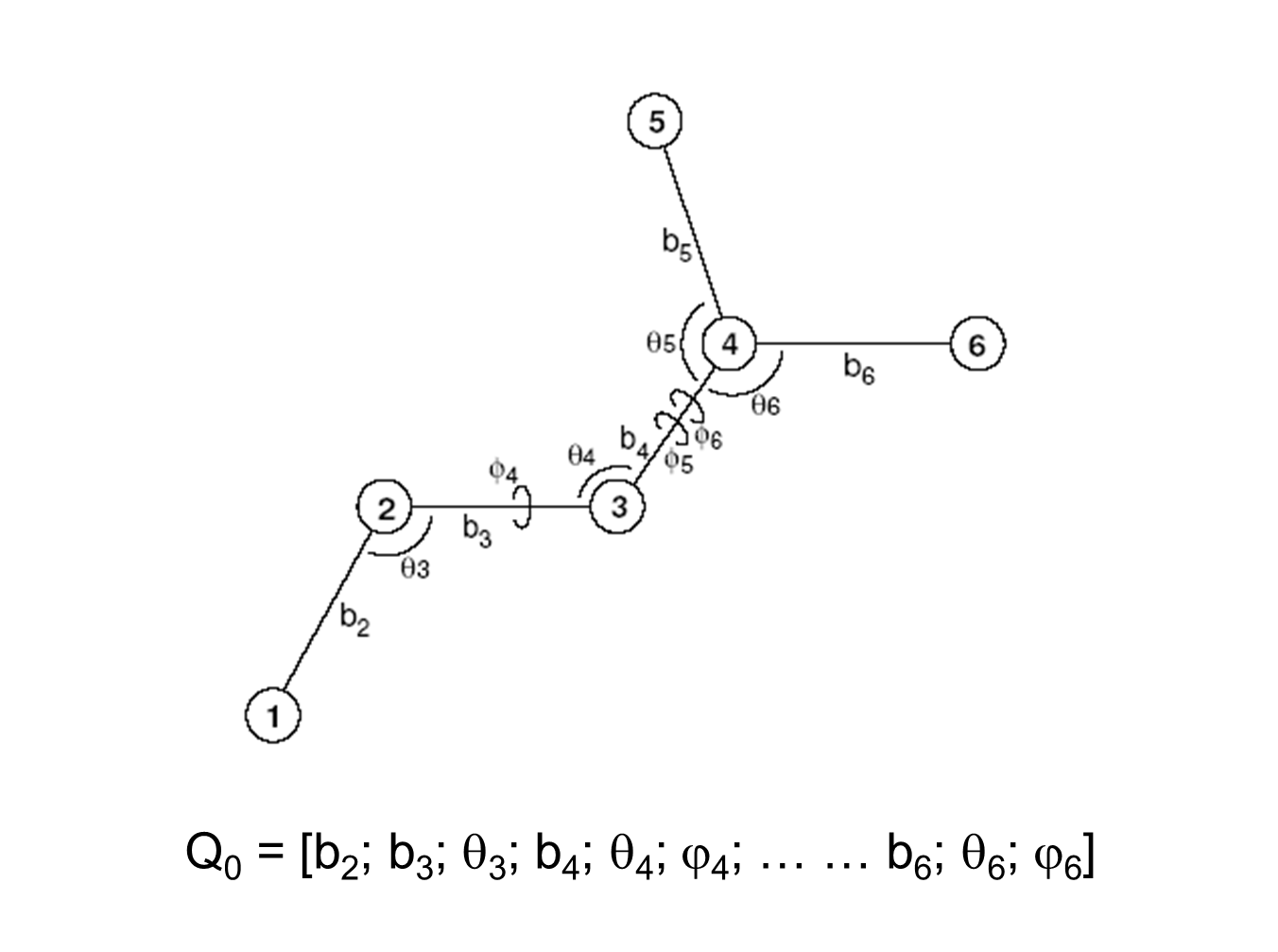
Internal Coordinate Molecular Dynamics: A Foundation for Multiscale Dynamics | The Journal of Physical Chemistry B
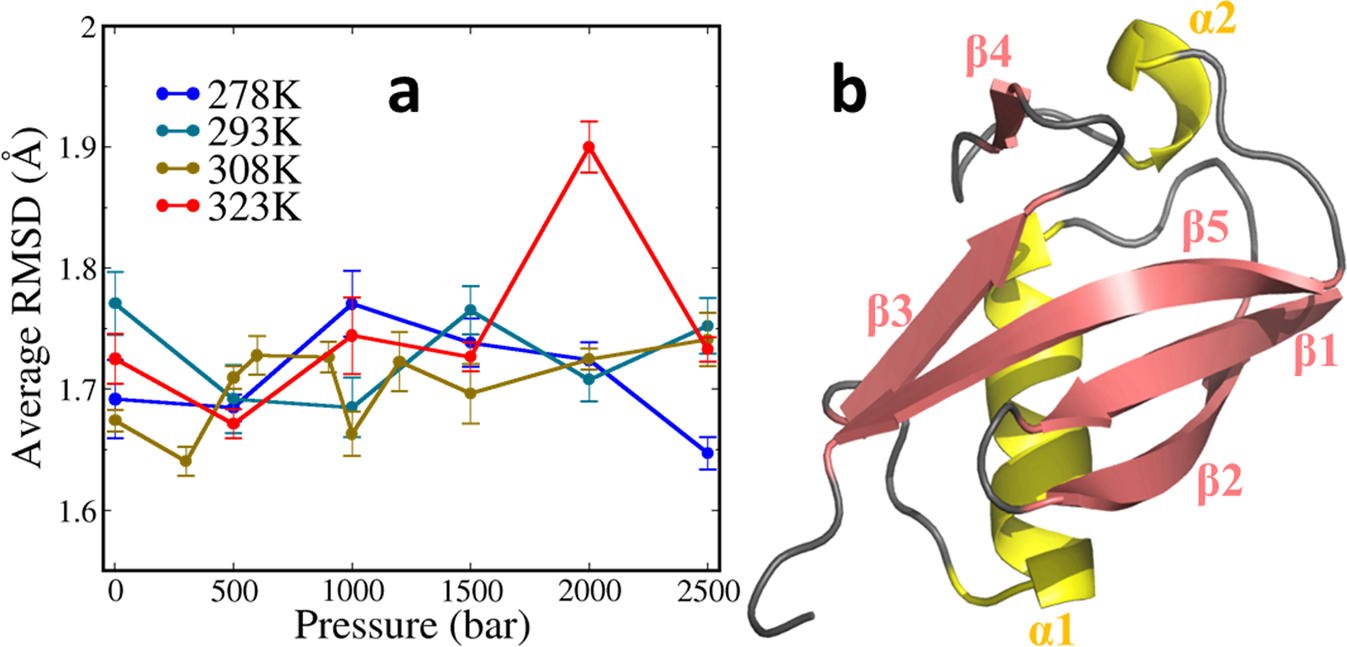
Validating the CHARMM36m protein force field with LJ-PME reveals altered hydrogen bonding dynamics under elevated pressures | Communications Chemistry

Branched polyethyleneimine: CHARMM force field and molecular dynamics simulations - Terteci‐Popescu - Journal of Computational Chemistry - Wiley Online Library

Computational Methods for Configurational Entropy Using Internal and Cartesian Coordinates | Journal of Chemical Theory and Computation

a) The internal potential plotted against the internal coordinate for... | Download Scientific Diagram

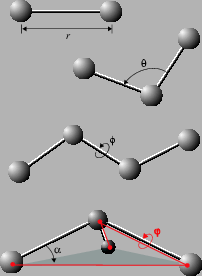
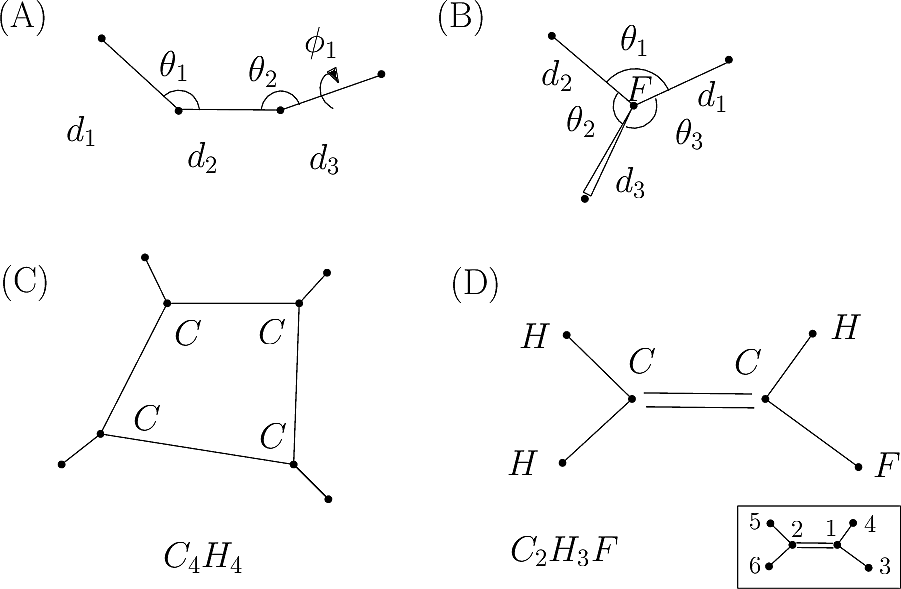
3 n - 6 internal bond-angle-torsion coordinates for a small molecule. | Download High-Quality Scientific Diagram

Spin‐projected QM/MM Free Energy Simulations for Oxidation Reaction of Guanine in B−DNA by Singlet Oxygen - Saito - 2021 - ChemPhysChem - Wiley Online Library